Introduction
Structure of DNA
The structure of DNA was first revealed through the extraction of nuclein by Meischer. Later, it was identified as nucleic acid, a polymer of nucleotides. These nucleotides are composed of phosphate, sugar, and nitrogenous bases. Watson and Crick’s model of DNA in 1953 demonstrated its double-helix form, a critical structure for heredity and genetic expression. DNA’s intricate design allows the encoding and transfer of genetic information essential for life.
Meischer extracted a substance from the nucleus of pus cells (leukocytes), which was responsible for heredity. He named that substance as NUCLEIN. Later on, Altmann studied the chemical nature of nuclein and confirmed that the substance is acidic in nature and that’s why he called nuclein as nucleic acid.
Nucleic acids are plolymers of nucleotides. And when we see the structure of a nucleotide, it is made from phosphate, sugar and nitrogenous bases.
Nucleotide = Phosphate + Ribose Sugar + Nitrogenous base
Nucleoside = Ribose Sugar + Nitrogenous base

Note: It is important to mention that there are two components of sugar: D-ribose and D-2’-deoxyribose
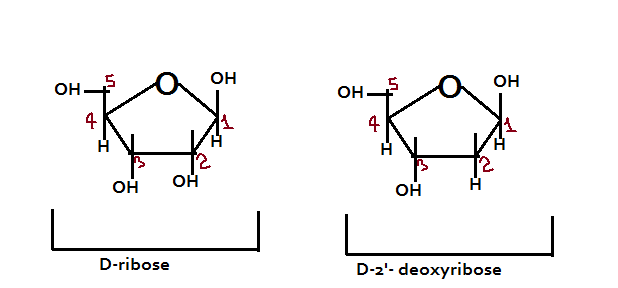
Structure of the Nitrogenous Bases:
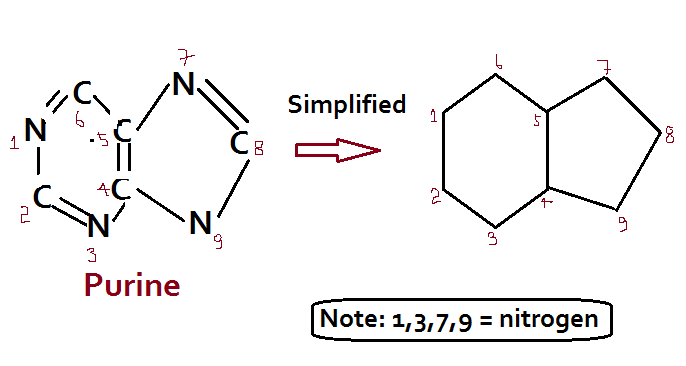
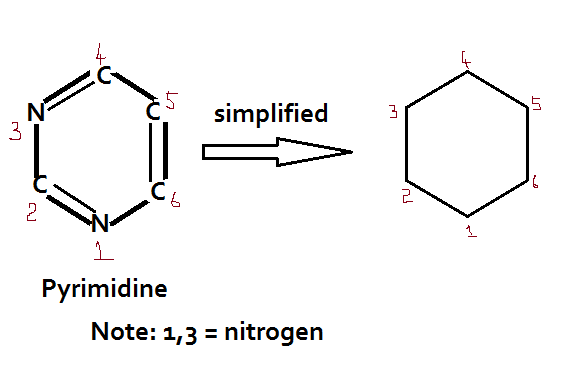
There are two groups of nitrogenous bases: Purine and Pyrimidines
Purine (Adenine – A; Guanine – G) = Double ring
Pyrimidine (Cytosine- C; Uracil – U; and Thymine — T) = Single ring Mnemonics: CUT
Attachment of bases in Nucleotides:
The nitrogenous bases are attached to the sugar moieties of nucleotides at the N-9 position of purines and the N-1 position of the pyrimidines. The bond formed between the sugar and nitrogenous base is the glycosidic bond in the beta configuration.
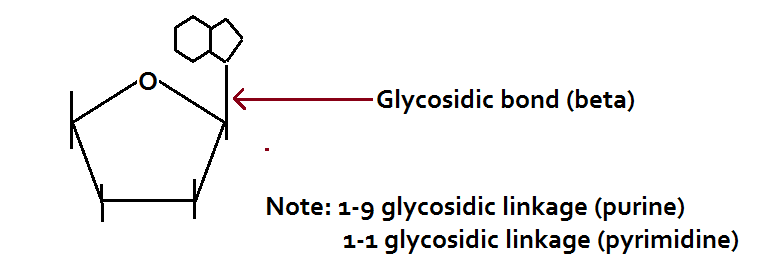
Nomenclature of Nitrogenous Bases

Polymerization of nucleotides:
When one nucleotide attaches with the other nucleotide, a bond is formed between the 3’-OH group of the first nucleotide sugar to the 5’-phosphate groups of the other nucleotide. The bond is shown below in the picture:
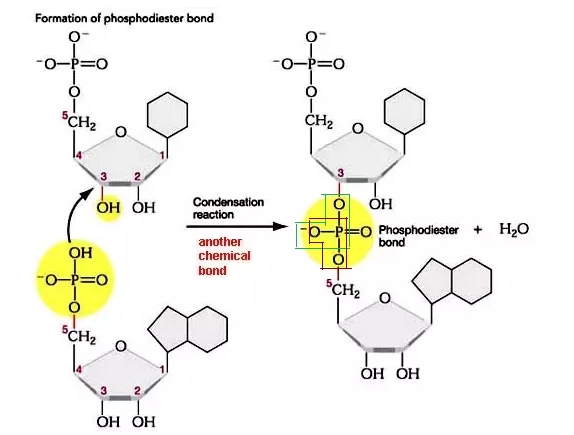
Q. How many water molecules will be released by the polymerization of 100 nucleic acids?
- 50
- 49
- 99
- 100
Ans: c
To find the number of molecules of water released due to polymerization of nucleic acid – use the formula (n-1), where n is the number of nucleic acids.
Structure of DNA
The three dimensional structure of DNA was proposed by Watson and Crick in 1953. However, the basis of this structure was given by three other scientists including Rosalind Franklin, Wilkins and Chargaff.
Rosalind Franklin:
She worked in the laboratory of Maurice Wilkins. She used X-ray diffraction to study wet fibers of DNA. She made marked advances in X-ray diffraction technique with DNA. The diffraction pattern she obtained suggested several structural features of DNA such as:

Franklin and Wilkins measured the width of the DNA helix as 2nm or 20 Angstrom. In each DNA helix turn, there are 10 layers of bases and the distance between each layer is 0.34 nm or 3.4 Angstrom; and thus for each complete turn – the length is 3.4 nm or 34 Angstrom. One complete helical turn is called PITCH.
Erwin Chargaff:
He pioneered many of the biochemical techniques for the isolation, purification and measurement of nucleic acids from living cells. It was known that DNA contained 4 nitrogenous bases: A, T, G and C. Chargaff analyzed the base composition of DNA isolated from many different species. The experimental observation of Chargaff revealed that:
% of adenine = % of thymine
% of cytosine = % of guanine
This observation is known as Chargaff’s rule. It is however, important to note that Chargaff principle is true only form double stranded DNA.
[A] + [G] = [T] + [C]
Or, [A+G] / [T+C] =1 (constant)
Total purine = total pyrimidine; however, [A] + [T] / [G+C] varies with organisms, but for each species, it is constant. For instance, humans = [A+T] / [G+C] = 1.55 i.e., greater than 1; and for E. coli – [A+T] / [G+C] = 0.43 i.e., less than 1.
In 1953, Watson and Crick gave the three dimensional model of DNA. According to the model:
- Two polynucleotide chains are present in DNA and the sugar-phosphate forms the backbone of DNA
- The two strands of DNA are in anti-parallel polarity
- Adenine joins with thymine by two hydrogen bonds and guanine joins with cytosine by three hydrogen bonds
- The DNA helix shows right handed coiling
The hallmark of DNA structure given by Watson and Crick is that A combines with T and G combines with C. And the base pairs are joined by hydrogen bonds. This finding was based on Chargaff’s DNA analysis. Watson, Crick and Wilkins received Nobel Prize for the discovery of DNA in 1962
DNA types:
| DNA | Helical diameter | Base pair / turn | Rotation | Vertical rise / bp |
| A | 23 Angstrom | 11 | RH | 2.56 Angstrom |
| B | 20 Angstrom | 10 | RH | 3.4 Angstrom |
| Z | 18.4 Angstrom | 12 | LH | 3.8 Angstrom |
| C | 19 Angstrom | 9.33 | RH | 3.3 Angstrom |
Salient Features of Double-helix structure of DNA:
- DNA is made up of two polynucleoide chains with sugar-phosphate as backbone and the bases project inside.
- The two strands of DNA have anti-parallel polarity. It means, if one chain has a polarity of 5 à 3, the other will have 3 à 5 polarity.
- The bases present inside the double helical strand are paired through hydrogen bonds. Adenine pairs with thymine with a double hydrogen bond; cytosine pairs with guanine with a triple hydrogen bond.
- The two strands of DNA are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm or 34 Angstrom.

Packaging of DNA Helix
We know that the distance between two consecutive base pairs as 0.34 nm or 3.4 Angstrom. With this information, we can calculate the length of DNA double helix in a mammalian cell. This can be done by simply multiplying the total number of base pairs with distance between the consecutive base pair. For example, the total number of base pairs in human DNA is 6.6 x 109 bp and the distance between each base pair is 0.34 nm; so the total length of DNA = 6.6 x 109 x 0.34 x 10-9 = 2.2 meter.
We know that the dimension of a typical nucleus is 10-6 meter. It is thus impossible to accommodate a DNA having a length of 2.2 meter, but it is true. DNA is very well accommodated in a nucleus. Now, you may ask the question – how is it possible? Well, the answer lies in DNA packaging. In eukaryotes, DNA is packaged in a very complex manner.
The two types of proteins required for DNA packaging include histone protein and non-histone chromosomal protein. Histone protein is of low molecular weight and basic in nature (possess positive charge). Non-histone chromosomal protein is of high molecular weight and acidic in nature (possess negative charge).
Histone proteins are rich in two amino acids i.e., lysine and arginine. Both lysine and arginine are basic amino acids, which are present in high concentration in the histone protein. On the other hand, non-histone proteins are rich in tyrosine and tryptophan.
There are five different kinds of histone proteins present such as: H1, H2A, H2B, H3 and H4. Out of these five histone proteins H1, H2A and H2B are rich in lysine and among these three H1 has the highest lysine concentration. H3 and H4 are rich in arginine.
The three non-histone proteins include structural / scaffold protein – helps in coiling of DNA. The second is enzymatic protein such as DNAP – DNA polymerase, RNAP – RNA polymerase. The third one is regulatory protein / HMG (high mobility group) – involved in gene expression. Higher level of coiling of DNA is assisted by non-histone proteins.
Q. How regulatory proteins help in gene expression?
A. DNA is negatively charged molecule and histone proteins are positively charged. Till the time histone protein is bound to the DNA – no gene expression will occur. However, when regulatory protein interacts with the histone protein – it helps in histone protein modification leading to the instability and thus removal of histone proteins. Once histone protein is removed from the DNA molecule – the gene will be expressed.
Mechanism of DNA Packaging:
In DNA histone proteins are present in two sets to form a structure, called Octamer / Nu body or Core molecule.
Out of five histone proteins, four of them are present in two sets to form an octamer. H2A, H2B, H3, and H4 in two sets form an octamer. The DNA molecule is wrapped around the octamer (1 full turn and ¾ turn). However, H1 protein is involved in the clipping of DNA. All these structures together form the NUCLEOSOME (DNA + histone octamer + H1 protein). So many nucleosomes would be formed to package DNA.
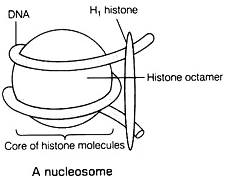
The diameter of nucleosome is 10 nm and the length of nucleosome is 6 nm. Each nucleosome consists of DNA having 200 bp.
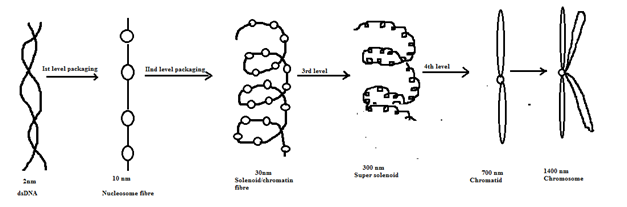
So many nucleosomes would be formed. After second level packaging, 30nm solenoid chromatin fibre would be formed. Again, the solenoid would coil i.e., 3rd level coiling to form super solenoid having a diameter of 300 nm. The fourth level coiling would lead to the formation of chromatid having a diameter of 700 nm. The chromatid after duplication would form chromosome with a diameter of 1400 nm.
Q. Which of the following is true for the two types of chromatin?
- Heterochromatin is darkly stained while euchromatin is lightly stained
- Heterochromatin is tightly packed while euchromatin is loosely packed
- Heterochromatin is transcriptionally inactive while euchromatin is transcriptionally active
- All of the above
Ans: d
Conclusion
In conclusion, the structure of DNA plays a fundamental role in the transmission of genetic information, as first outlined by Watson and Crick’s model in 1953. The double-helix structure of DNA ensures the precise encoding and replication of genetic material, which is vital for life. Understanding the structure of DNA continues to be essential in fields such as genetics, medicine, and biotechnology, providing insights into heredity and disease mechanisms.

