Introduction to Transcription in Prokaryotes
The process of copying genetic information from one strand of DNA into RNA is called transcription – it is the first step of the expression of genes into proteins. In other words, transcription is a process in which one strand of DNA is converted into RNA. However, in RNA, the nitrogen base, Thymine (T) is replaced by Uracil (U) and deoxyribose sugar in DNA is replaced by ribose sugar in RNA.
The RNA formed from DNA through transcription is called a TRANSCRIPT. It is important to mention that in transcription only one of the DNA strands is copied into RNA and transcription takes place only in 5’ — > 3’ direction. The question you may ask – why both DNA strands are not copied during transcription? Well, there are two simple reasons:
(i) If both DNA strands would form RNA, the newly formed RNAs would be complementary to each other and would form a double stranded RNA. This would prevent RNA from being transcribed into protein.
(ii) If both DNA strands would code for RNA molecules – the two RNA strands would have different sequences. And if they code for proteins – the sequence of amino acids in the newly formed proteins would be different. This would complicate the genetic information transfer machinery.

Differences in the Process of Transcription in Prokaryotes versus Eukaryotes
- In prokaryotes (E. coli), transcription and translation occurs in the cytoplasm. However, in eukaryotes, transcription occurs in the nucleus and translation occurs in the cytoplasm.
- Prokaryotic DNA is easily accessible to RNA polymerase in prokaryotes as compared to eukaryotes. In case of eukaryotes, DNA is highly coiled and packed by histone proteins.
- The mRNA produced in prokaryotes is not modified further, but in eukaryotic cells, the mRNA formed is further modified by RNA splicing, 5’-end capping and 3’-end tailing.
Transcription Unit in DNA:
It is the segment of DNA that is to be transcribed. In DNA, many transcription units can be found. A transcription unit consists of:
- Promoter
- The structural gene
- A terminator
Q. DNA is double stranded, which DNA strand would be transcribed?
Well, we know that transcription like DNA replication takes place only in 5’ — > 3’ direction, the DNA strand having a polarity of 3’ — > 5’ will act as a template – known as NON-CODING STRAND or NON SENSE STRAND; the other strand with a polarity of 5’ — > 3’ will be called as a CODING STRAND or SENSE STRAND because the new RNA that would form have the same sequence as the coding strand.
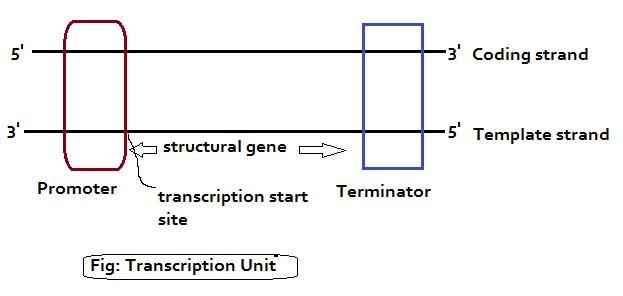
Promoter: It is located towards the 5’-end (upstream) of the structural gene with reference to the coding strand (5’—3’ DNA strand). Promoter is a DNA sequence that provides binding site for RNA polymerase enzyme. And it is the promoter that defines the template and coding strands. In other words, the DNA strand consisting of a promoter sequence is called as CODING STRAND. The bases in the promoter binding site are numbered in relation to the transcription start site.

The conventional numbering system of promoters
Most of the promoter regions are labeled with negative numbers. Bases preceding the start site are numbered in a negative direction; there is no base numbered zero. And bases to the right are numbered in a positive direction.
Structural Gene Present in the Transcription Unit:
A gene simply is defined as the functional unit of inheritance. A gene (DNA segment) can form mRNA, rRNA and tRNA; however, there is a special term used for mRNA formation that eventually forms protein. This special term is called as CISTRON. Gene, therefore, is a broader term and cistron is a narrow term even though both are coding DNA fragments. In other words, when we say cistron, it refers to a segment of DNA that code for a polypeptide. Different genes are responsible for forming different kinds of RNAs. A single gene cannot form all RNAs.
Note: In eukaryotes, since genes are separated and not present in tandem, only MONOCISTRONIC CONDITION is present. On the other hand, in prokaryotes, structural gene (a combination of genes in tandem) code for more than one polypeptide, called POLYCISTRONIC CONDITION. Besides, in eukaryotes, coding sequences are interrupted by non-coding sequence, the coding DNA sequence is called EXON and the non-coding sequence is called INTRON. In case of prokaryotes, introns are absent except in archaebacteria.
Types of RNA and the Process of Transcription
In bacteria, three types of RNAs are present i.e., mRNA, rRNA, and tRNA. All three RNAs are required for protein synthesis. For example, mRNA acts as a template; tRNA is responsible for bringing amino acids and reads the genetic code; rRNA plays structural and catalytic role during translation.
Note: In prokaryotes, there is a single dna-dependent RNA polymerase that catalyzes transcription of all types of RNA.
Structure of RNA polymerase Enzyme:
The enzyme consists of two parts – core enzyme and sigma factor. Both core enzyme and sigma factor together are called HOLOENZYME.
Core enzyme = α2ββ’ω
Sigma factor = σ

- Alpha subunit is required for core protein assembly. It also plays a role in promoter recognition. And assembly of beta and beta prime.
- Beta subunit is involved in catalysis, chain initiation and elongation. It means, it binds incoming nucleotides to form a polynucleotide.
- Beta prime binds to the DNA template
- Omega – required restoring denatured RNA polymerase in vitro to its fully functional form.
- Sigma factor – initiates transcription i.e., formation of 8-9 bp of RNA; sigma factor also recognizes -10 and -25 regions of the promoter.
Steps of Transcription:
1. Initiation
2. Elongation
3. Termination
Initiation:
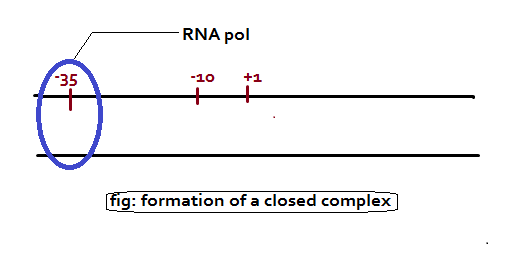
- Bacterial RNA polymerase (RNAP) that scans the DNA looking for a promoter sequence is called the RNA Polymerase Holoenzyme. RNAP is bound to approximately 60 base pairs.
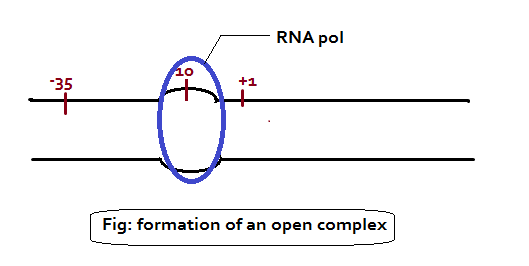
- The sigma unit binds to the -10 (PRIBNOW BOX – TATAAT) and -35 regions thus positioning the holoenzyme to initiate transcription correctly at the start site.
- The sigma subunit also has the role in separating (melting) the DNA strands (open complex formation) around the -10 region so that the core enzyme can bind tightly to the DNA in preparation for RNA synthesis.
- Once the core enzyme is bound, transcription begins and the sigma subunit dissociates from the rest of the complex.
Once the promoter region is detected by the holoenzyme, the sigma factor opens up the DNA helix for the core enzyme to initiate transcription. And then the sigma factor is released. It is important to note that transcription starts at +1 point, which lies before the protein-coding segment of the gene (usually at the sequence ATG), which is where translation usually starts. Hence the RNA which formed before the ATG sequence is called 5’-untranslated region or 5’ UTR.
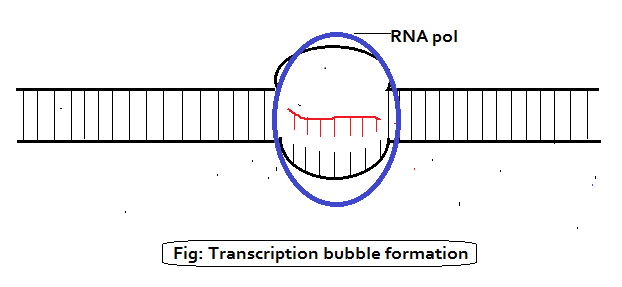
Elongation:
As the RNA polymerase moves along the DNA, it unwinds the DNA ahead of it and rewinds the DNA that has already been transcribed, in this way it maintains a region of single stranded DNA, called a TRANCRIPTION BUBBLE, within which the template strand is exposed. In the bubble, polymerase monitors the binding of a free ribonucleoside triphosphate to the next exposed base on the DNA template, and if there is a complementary match, adds it to the chain. The energy for the addition of a nucleotide is derived from splitting the high-energy triphosphate and releasing inorganic diphosphate, as follows:
NTP + (NMP)n ————— > (NMP)n+1 + PPi
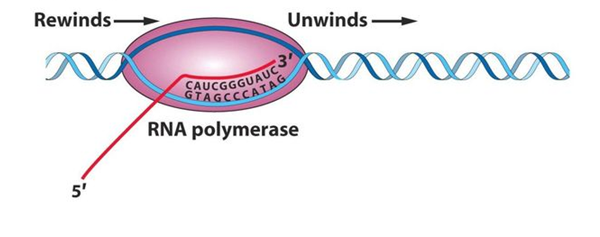
Elongation is performed by the core enzyme. After the initiation process, the core enzyme moves on the DNA template strand for RNA synthesis. The core enzyme will unwind the DNA strands to separate template from the non template strand. After unwinding of the helix, RNA synthesis will start leading to the formation of DNA-RNA hybrid. Like DNA replication, here also the substrate is activated from ribonucleoside monophosphate to ribonucleoside triphosphate. The RNA polymerase will continue moving on the DNA template to form RNA till it reaches the termination region.
Termination:
The transcription of an individual gene is terminated beyond the protein-coding segment of the gene, creating a 3’ untranslated region (3’ UTR) at the end of the transcript. Elongation continues until RNA polymerase recognizes special nucleotide sequences that act as a signal for chain termination. The encounter with the signal nucleotides initiates the release of the nascent RNA and the enzyme from the template. The two major mechanisms for the termination in E. coli are INTRINSIC and rho DEPENDENT.
- Intrinsic (rho independent)
- Rho dependent
Intrinsic method of termination:
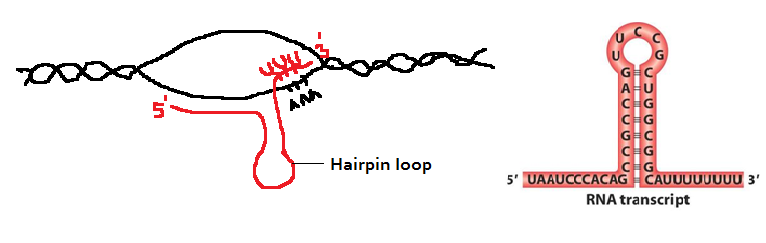
In this method, the termination is direct. The terminator sequence contains about 40bp, ending in a G-C rich stretch followed by a run of six or more A’s on the DNA template strand. The RNA strand opposite to G-C rich DNA template will also have GC rich RNA strand, which will form during transcription. These C and G bases are able to form complimentary hydrogen bonds with each other resulting in a hairpin loop. The loop is followed by a run of about eight U’s that correspond to the A residues on the DNA template.
Note: Normally, in the course of transcription elongation, RNA polymerase will pause if the short DNA-RNA hybrid in the transcription bubble is weak and will backtrack to stabilize the hybrid. For example, in the hairpins, the strength of the hybrid is determined by the relative number of G-C compared to A-U (or A-T in the RNA-DNA hybrid) base pairs.
In the intrinsic mechanism, the polymerase is believed to pause after synthesizing the U’s (which form a weak DNA-RNA hybrid). However, the backtracking polymerase encounters the hairpin loop which prevents it from finding a stable hybrid. This roadblock sets off the release of RNA from the polymerase and the release of the polymerase from the DNA template.
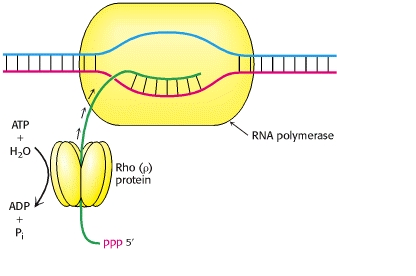
Rho Dependent Transcription:
In this method, the termination site is rich in C-residues and poor in G-residues. There is an upstream part, called rut (rho utilization) site at the 5’-end. Rho protein is a hexamer consisting of 6-identical subunits that bind to a nascent RNA chain at the rut site. These sites are located just upstream from sequences at which the RNA pol tends to pause.
The rho protein moves or follows the RNA pol on the RNA strand. The rho protein utilizes ATP to move on the RNA strand. As the RNA pol stops because of the termination bases, the rho protein collides with the enzyme and thus RNA pol releases from the RNA strand.