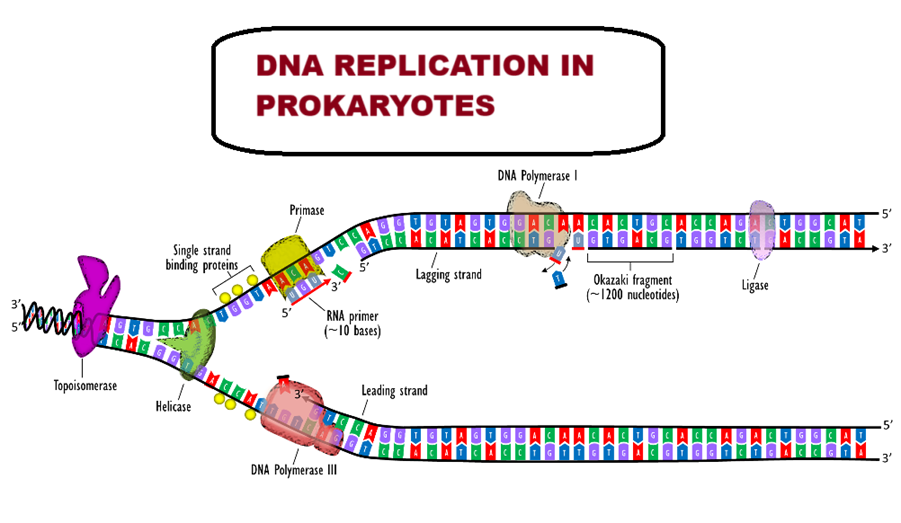
DNA replication in prokaryotes is completely understood as against DNA replication in eukaryotes because in the latter case, the process is very complex. That is the reason why we study in detail the DNA replication in prokaryotes. The organism E. coli (bacteria) is chosen to study the DNA replication in prokaryotes. Now, let’s understand the mechanism of DNA replication steps.
DNA replication starts at a specific region, called the OriC (Origin of Replication of Chromosome). It is a 245 base pair long sequence rich in AT nitrogen bases. Due to the presence of AT rich region, the unwinding becomes easier as A, and T are joined by only two weak H-bonds. On the other hand, G and C are joined by three H-bonds. It is important to mention that DNA replication in both prokaryotes and eukaryotes is bidirectional i.e., 5’ — > 3’ direction. We will discuss DNA replication in prokaryotes in different steps to make it easier to understand.
Steps of DNA Replication in Prokaryotes
- Unwinding of DNA double helix
- Addition of Primer to the DNA helix
- Polymerization of nucleotides for the formation of new DNA strands
- Proofreading Activity of DNA Polymerase
Unwinding of DNA Double Helix:
Unwinding of DNA is required to start DNA replication steps. This step involves different kinds of enzymes and proteins such as helicase, SSB protein (helix destabilizing protein) and topoisomerase enzyme. We’ll discuss the role of each enzyme and protein in detail.
Helicase enzyme is involved in the breaking down of hydrogen bonds between two strands of DNA. Unwinding of DNA involves expenditure of energy in the form of ATP. For example, one ATP is consumed for the breakage of one hydrogen bond (H-bond).
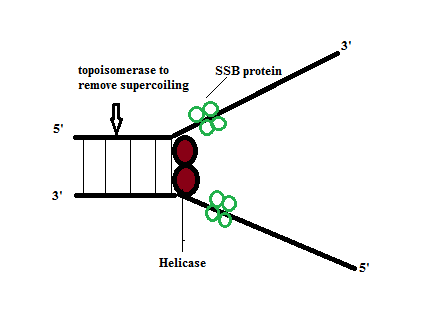
SSB Protein (Single Strand Binding Protein): It is also known as helix destabilizing protein that prevents recoiling of two DNA strands after the unwinding of DNA. In other words, SSB protein is present in the form of a TETRAMER that binds to each DNA helix to keep the helix unwounded.
Topoisomerase enzyme: When strand is being opened on one side, the coiling on the other side will increase, which is called SUPERCOILING. Due to supercoiling, opening of the helix would be difficult by the helicase enzyme. In order to make the process easier, the supercoiling needs to be removed and this is done by the enzyme – TOPOISOMERASE enzyme. There are two types of supercoiling – positive and negative supercoiling. Positive supercoiling means the strands are more twisted than the normal. And the negative supercoiling means the strands are less twisted than the normal twist. However, topoisomerase activity fixes both the problems. In case of positive supercoiling – the topoisomerase has two weapons – one to cut one strand of DNA and relieve the tension and the other weapon is ligase to again attach the cut DNA. In prokaryotes, GYRASE is present and it functions as both of helicase and topoisomerase.
Addition of Primer to the DNA helix:
Primer is a short RNA oligonucleotide with free 3’-OH group, which is the site for DNA polymerization. The primer is complementary to the DNA template sequence. Primer is synthesized by the enzyme PRIMASE.
Polymerization of nucleotides for the formation of new DNA strands:
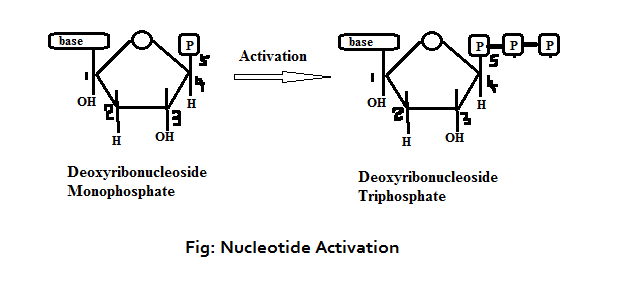
The raw nucleotides (deoxyribonucleotide or deoxyribonucleoside mono phosphate) present in the nucleus will undergo activation where two more phosphates will be added leading to the formation of deoxyribonucleoside triphosphate. In other words, 4 different types of nucleotides would be formed like deoxy adenosine triphosphate (deATP), deoxy cytidine triphosphate (deCTP), deoxy guanosine triphosphate (deGTP) and deoxy thymidine triphosphate (de TTP). These activated nucleotides would act like substrate for DNA polymerization. A phosphodiester bond is formed between the 3’-OH group of primer and the 5’-phosphate group of the deoxyribonucleotides.
Enzyme: DNA polymerase is involved in DNA replication in prokaryotes. There are three different types of DNA polymerases:
- DNA polymerase I — also known as Kornberg enzyme, named after the scientist who discovered it. This enzyme is composed of a single polypeptide. It is responsible for removing RNA primers and replaces them with DNA. It also possesses 3’à 5’ exonuclease and 5’ à 3’ exonuclease for proofreading.
- DNA polymerase II – this enzyme is responsible for polymerization of DNA in the 5’ à 3’ direction. However, its polymerization rate is very slow i.e., 50 nucleotides per minute. This enzyme also contains 3’ à 5’ exonuclease.
- DNA polymerase III – it is the main DNA polymerization enzyme having a great polymerization speed of 2000 bp per second. This enzyme also contains 3’ à 5’ exonuclease.
Summary of DNA polymerase enzyme:
| Enzyme | DNA pol I | DNA pol II | DNA pol III |
| 5’– 3’ polymerase | Yes | Yes | Yes |
| 3’ — 5’ exonuclease | Yes | Yes | Yes |
| 5’ — 3’ exonuclease | Yes | No | No |
| Polymerization rate | 16-20 | 40 | 250-1000 |
Formation of Okazaki Fragments:
During DNA replication one template replicates a continuous strand called leading strand and the other forms a discontinuous strand, called the lagging strand. The lagging strand is also known as Okazaki fragments. Okazaki fragments are joined by DNA ligase activity. The leading strand is formed towards the replication fork and lagging strand is formed away from the replication fork. The leading strand requires only one primer while the lagging strand requires many primers.
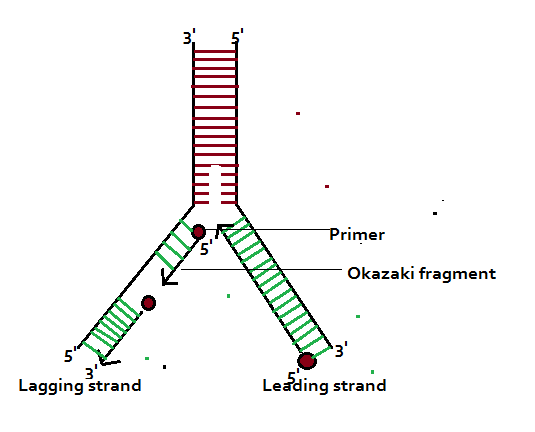
As DNA replication initiates on the DNA template strand in the 5’ à 3’ direction – a continuous new strand is formed on the 3’—5’ DNA template. This new strand is called LEADING DAUGHTER STRAND. On the other hand, another daughter strand is formed on another DNA template, but this formation is not continuous rather in the form of short pieces. This new strand in pieces is called LAGGING DAUGHTER STRAND. And these short pieces of DNA are called OKAZAKI FRAGMENTS and these segments are about 1,000 – 2,000 nucleotides long in prokaryotes.
Proofreading Activity of DNA Polymerase
5’ — > 3’ exonuclease is required for the removal of DNA sequence from the 5’-end. It means, this enzyme activity is important for the removal of primer. This activity is shown by DNA pol I only and this DNA pol is also responsible for substituting primer with nucleotides. On the other hand, 3’ — > 5’ exonuclease activity is required for proofreading and thus present in all three DNA polymerases.
Note: the main polymerizing activity is shown by DNA pol III. Besides, Pol I is a major repair enzyme while pol II is a minor repair enzyme. The gap formed in lagging strand is filled by DNA pol I and attached by DNA ligase activity.
Frequently Asked Questions (FAQs)
Q. Are DNA gyrase and topoisomerase the same enzyme?
A. Yes, they are related. DNA gyrase is a specific form of topoisomerase, known as topoisomerase II. It functions by cutting both strands of DNA, relaxing the supercoils, and then rejoining the strands.
Q. If DNA polymerization only proceeds when bases pair correctly, how can DNA polymerase still make mistakes?
A. Even though DNA polymerase usually adds the correct nucleotide only when it matches the template base, errors can still occur because the process isn’t perfect. During replication, especially on the lagging strand where DNA is formed in short Okazaki fragments, mismatches may occasionally slip through before the enzyme detects them. DNA polymerases have a 3′→5′ exonuclease activity that proofreads and removes these incorrect bases, but if an error escapes this proofreading step, it becomes a mutation.
Q. In which direction does DNA synthesis occur — 5′ to 3′ or the reverse? And what are the orientations of the leading and lagging strands?
A. DNA synthesis always occurs in the 5′ to 3′ direction, meaning new nucleotides are added to the free 3′-OH end of the growing strand. The leading strand is synthesized continuously in the same direction as the replication fork movement (toward the fork), while the lagging strand is synthesized discontinuously in short fragments called Okazaki fragments, which form in the direction opposite to the fork movement.
Q. What function do SSB proteins serve?
A. SSB proteins, also known as single-strand binding proteins or helix-destabilizing proteins, bind to the separated DNA strands during replication to keep them from reannealing. They stabilize the unwound DNA, ensuring that the strands remain open and accessible for replication to continue smoothly.
Q. Since eukaryotic chromosomes are linear, DNA is lost from their ends (telomeres) with each replication. Does this loss occur every time, and can it be restored, or does it impact other cellular processes?
A. Yes. A small portion of DNA is lost from chromosome ends (telomeres) during each replication because the lagging strand cannot be fully copied. Most cells do not recover this loss since telomerase is inactive, leading to gradual telomere shortening, which eventually causes cell aging or death.
Q. If the genome represents the haploid DNA content of a cell, how does it contain 3 billion base pairs — or does that number actually describe the diploid DNA content?
A. The human genome refers to the haploid set of DNA, meaning one complete set of chromosomes. This set contains about 3 billion base pairs. In diploid cells, which have two sets of chromosomes (one from each parent), the total doubles to about 6 billion base pairs.